A computer-based SimBio exploration of evolution
 |
This tutorial-style
inquiry contextualizes abstract evolutionary
genetics concepts to a classic case of how malaria
influences the prevalence of sickle-cell anemia in
humans. This is done using a guided
inquiry-based computer simulation which allows
students develop hypotheses and test predictions
regarding the following questions:
|
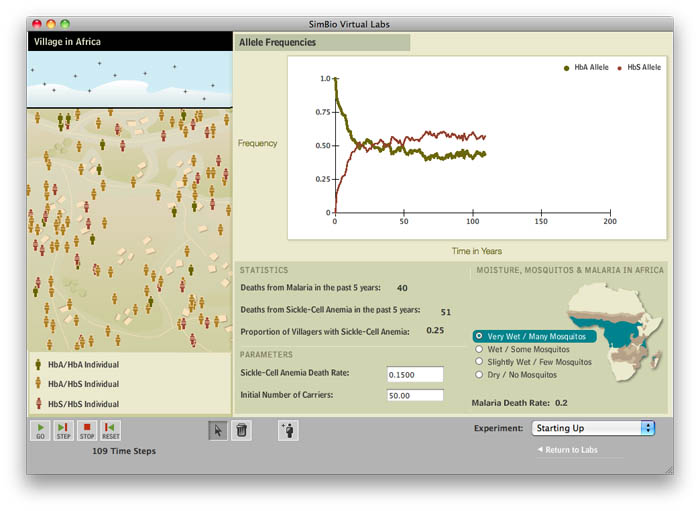
In this lab students practice using the Hardy-Weinberg equation to calculate the expected proportion of sickle-cell carriers from HbS and HbA allele frequencies. Then they examine how and why allele and genotype frequencies change with changes in malaria risk and also under different "founder" scenarios. Finally they explore genetic drift without selection by looking at different-sized villages where sickle cell anemia has been cured and malaria eradicated. The lab is also an excellent follow-up to our case-study based lab which explores the genetics and molecular biology of sickle cell anemia. A preview of the computer simulation are available through SimBio. Our adaption of the student instructions for this lab is more hypothesis-driven, and we have developed case studies to support and contextualize the lab simulation when it is taught as a stand-alone lab.
Conceptual Learning Objectives - Upon completion of this lab, students should be able to
- use the Hardy Weinberg equations to calculate genotype frequencies from allele frequencies and vice versa.
- explain the utility of using the Hardy-Weinberg model to test hypotheses about the role that natural selection, mutations, and population size (genetic drift) play in the evolution of populations.
- explain how environmental conditions can influence the fitness of different alleles or genotypes and therefore their relative prevalence in populations.
- predict how the fitness coefficients of each genotype in the sickle cell-malaria system will change under different climate conditions in Africa, and can explain why these different fitness coefficients produce varied genotypic frequencies among the 3 genotypes.
- explain how population size (genetic drift) influences the allele frequencies over time both when there is selection for an allele (or genotype) and when selection is absent.
- discuss how evidence from the simulation exemplifies a case of human evolution through natural selection.
- discuss how environmental conditions and selection pressures influence the initial occurrence and establishment of a new allele in a population.
Scientific Skills - In this lab students practice and receive feedback on
- identifying and interpreting trends in a line graph that depicting the changes in allele frequencies over time.
- understanding that mathematical models of natural phenomena are built from theories and have underlying assumptions.
- applying a mathematical model (Hardy-Weinberg equation) to generate estimates of related parameters (i.e. estimating genotypic frequencies from graphically displayed allele frequencies).
- using biological concepts to formulate and justify data predictions which emerge from a scientific hypothesis.
- interpreting and analyzing data trends presented both in graphical and table form to evaluate a hypothesis.
Required Materials
- SimBio Simulation
- Student team computers